The general procedure is as follows:
- Build a model for the training data set.
- Evaluate model performance on the test data set.
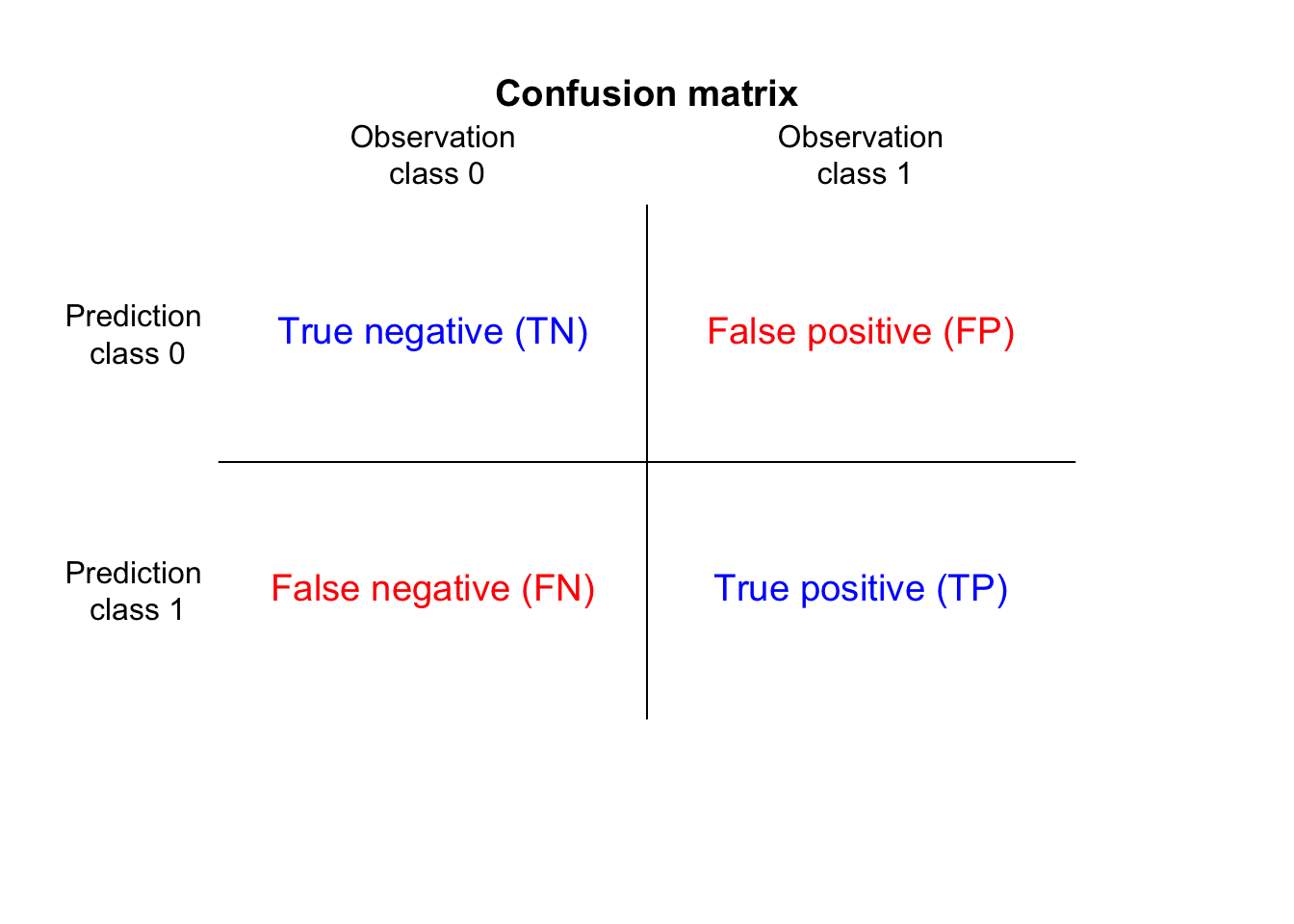
We evaluate the model performance by calculating the model accuracy, the confusion matrix and Cohen’s kappa. The confusion matrix is a specific table that visualizes the performance of a predictive model. Each column of the matrix represents the instances in a actual class while each row represents the instances in an predicted class (or sometimes vice versa).
Cohen’s kappa coefficient, $κ$, is a statistic which measures inter-rater agreement for categorical items. It takes a value of $κ=0$ if the items agreement is due to chance. A value of $κ=1$ indicates a perfect model fit.
The definition of $κ$ is:
$$\kappa=\frac{p_0-p_e}{1-p_e},$$where $p_0$ is the relative observed agreement (identical to accuracy), and $p_e$ is the hypothetical probability of chance agreement, using the observed data to calculate the probabilities of each observer randomly saying each category.
The baseline model¶
In the baseline model we predict the response variable HOT_DAY by including all 15 features in
the data set. We use the $L_2$-regularized logistic regression model
(ridge regression:
alpha = 0) provided by the sklearn package. In particular we apply the LinearRegressionCV
class, which performs by default
cross-validation) on
the data set and evaluates the model performance.
