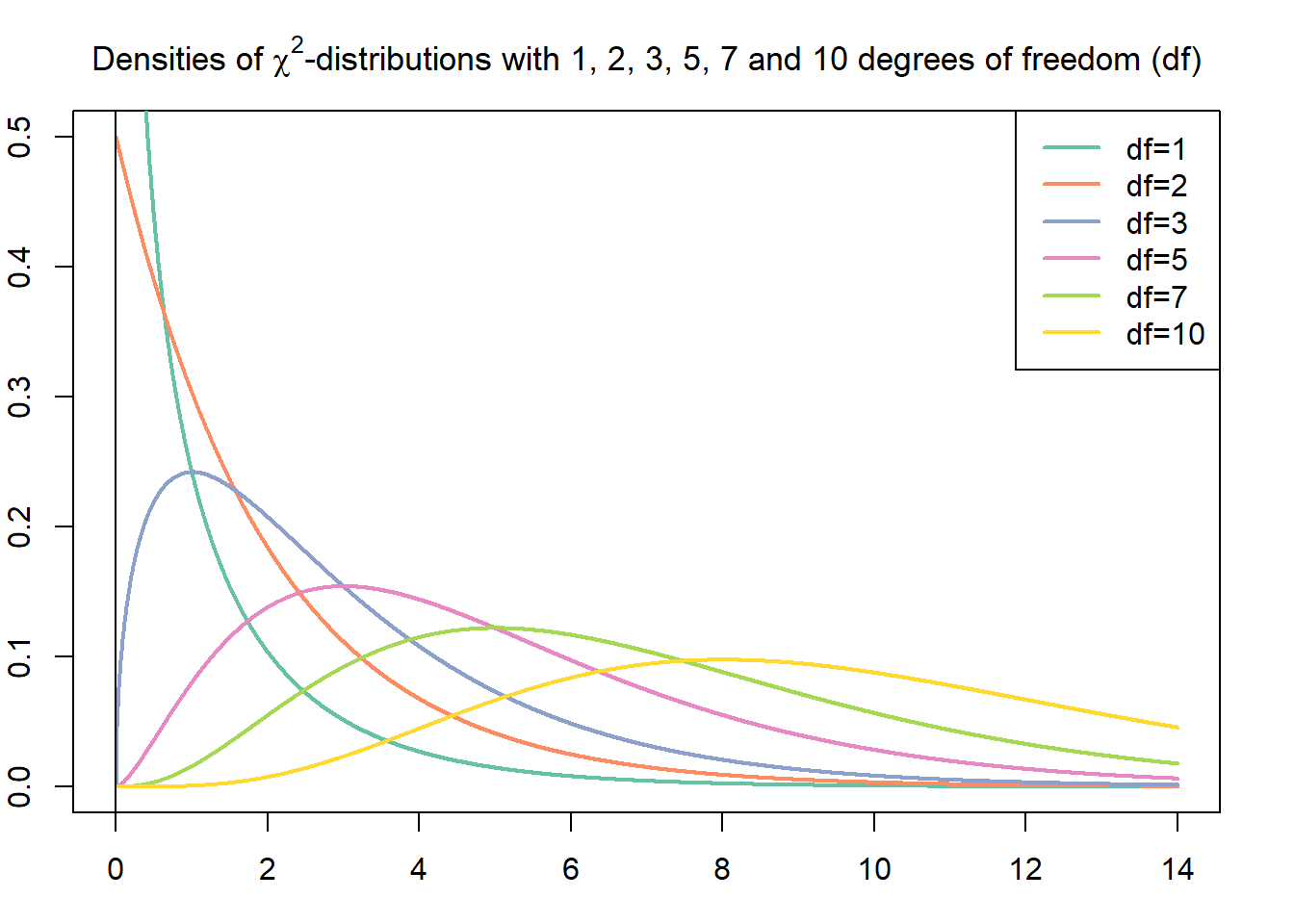
Inferences for one population standard deviation are based on the *chi-square ($\chi^2$) distribution*. A $\chi^{2}$-distribution is a right-skewed probability density curve. The shape of the $\chi^{2}$-curve is determined by its degrees of freedom $(df)$.
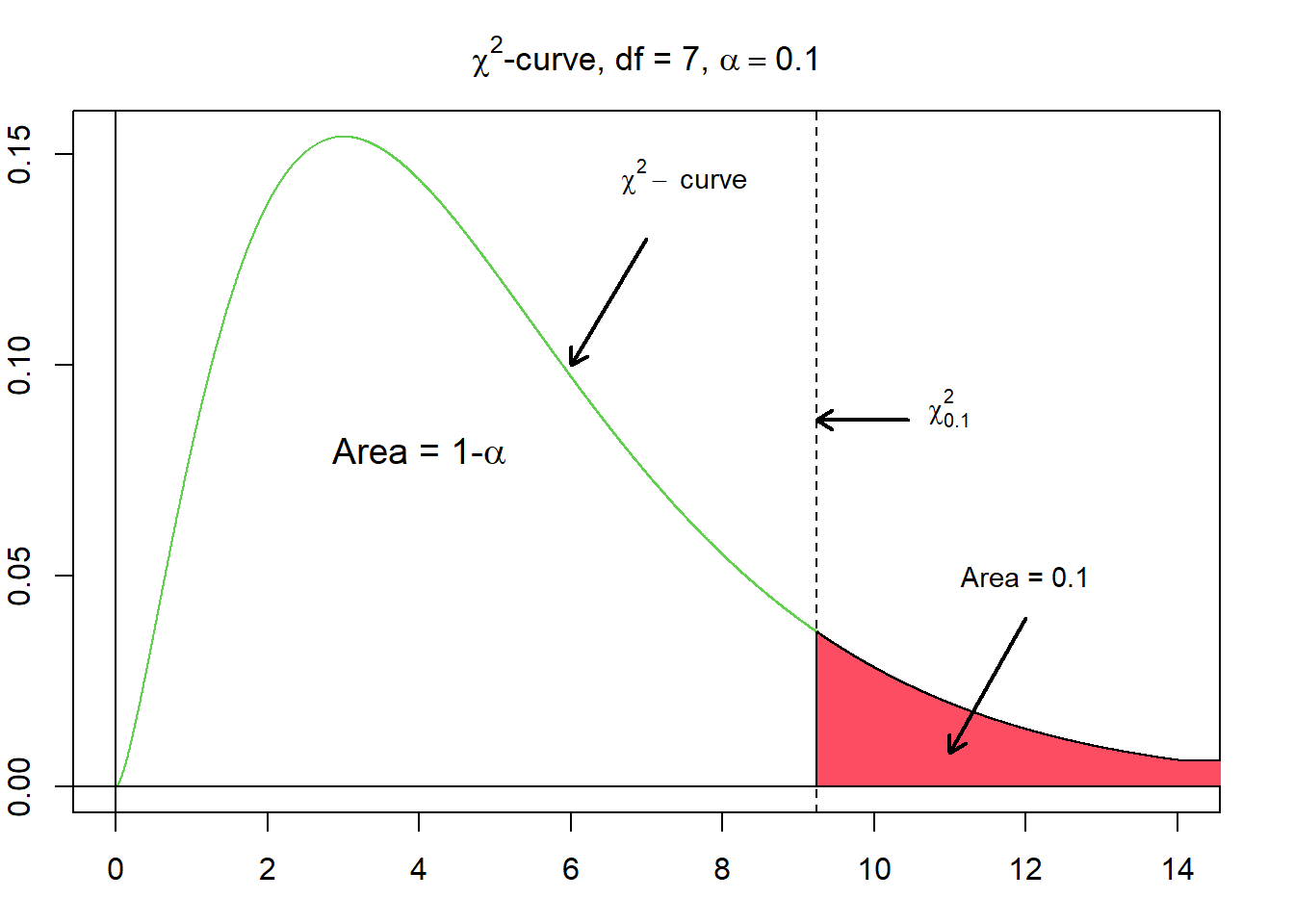
In order to perform a hypothesis test for one population standard deviation, we relate a $\chi^{2}$-value to a specified area under a $\chi^{2}$-curve. Either we consult a $\chi^{2}$-table to look up that value, or we use Python directly and the corresponding package universe for those purposes.
Given $\alpha$, where $\alpha$ corresponds to a probability between 0 and 1, $\chi^{2}_{\alpha}$ denotes the $\chi^{2}$-value having the area $\alpha$ to its right under a $\chi^{2}$-curve.
Interval Estimation of $\sigma$¶
The $100(1 − \alpha)\ \%$ confidence interval for $\sigma$ is:
$$\sqrt { \frac {n - 1} {\chi^2_{\alpha / 2}}} \le \sigma \le \sqrt { \frac{n - 1} {\chi^2_{1 - \alpha / 2}} }\text{,}$$where $n$ is the sample size of the sample data.
One standard deviation $\chi^{2}$-test¶
The hypothesis testing procedure for one standard deviation is called one standard deviation $\chi^{2}$-test. Hypothesis testing for variances follows the same step-wise procedure as hypothesis tests for the mean:
- State the null hypothesis $H_{0}$ and alternative hypothesis $H_{A}$.
- Decide on the significance level, $\alpha$.
- Compute the value of the test statistic.
- Determine the p-value.
- If $p \le \alpha$, reject $H_{0}$; otherwise, do not reject $H_{0}$.
- Interpret the result of the hypothesis test.
The test statistic for a hypothesis test with the null hypothesis $H_{0}: \,\sigma = \sigma_{0}$ for a normally distributed variable is given by:
$$\chi^{2} = \frac {n - 1} {\sigma^{2}_{0}} s^{2} \text{.}$$The variable follows a $\chi^{2}$-distribution with $n - 1$ degrees of freedom.
Be aware, that the one standard deviation $\chi^{2}$-test is not robust against violations of the normality assumption (Weiss, 2010).
